The Missing Link: How to Combine Neuroimaging Data with Computational Models of Behavior
This course will take place on 9 June 2019 from 13:00 to 16:30 at the OHBM meeting in Rome.
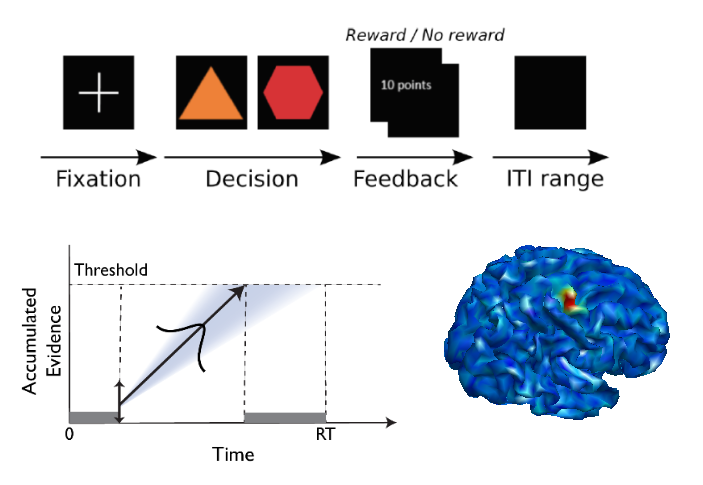
Cognitive computational models proved useful for the scientific community in a wide range of research areas, such as decision making, learning and memory. Modelling behaviour offers a way to see beyond raw behavioral measures, such as accuracies and reaction times, and estimate latent cognitive parameters which drive behaviour. The key challenge of the field lies in integrating cognitive models of behaviour with the rich ensemble of neural data. Neuroimaging technologies offer a plethora of ways to sample neural data, each having its benefits and drawbacks.
In this workshop, we aim to bridge these two well-developed and self-proved areas.
If you are a researcher working with a variety of neuroimaging methods, and incorporate behavioral measures in your experiments, this course could be beneficiary for you.
We will be taking questions during the course on OHBM app. If you have any question for the presenters, please choose this course on the website and type your questions!
Course material will be available here soon.
Program
13:00 - 13:20 Wojciech Zajkowski “Defining the missing link: introduction to combining neural data with cognitive models of behavior.”
Abstract
In recent years, the amount of neuroimaging data we collect has been growing in an exponential fashion. Due to this, the issue of developing methods that aid in drawing meaningful conclusions from large and rich (often multimodal) datasets has gained paramount importance. This talk will consist of three segments. I will start by providing an overview of the purpose of using cognitive models of behavior and how can they be utilized together with brain data, using models such as Drift-Diffusion Model or Linear Ballistic Accumulator as examples. I will then explain the nature of the linking problem. Using the Marr’s levels of analysis framework, I will go through the rationale behind why is it crucial for our understanding of brain and behavior. Finally, I will introduce different methods of dealing with the problem: from a simple correlation to recently developed frameworks, such as joint modelling, which utilize the covariability of neural and behavioural data to draw meaningful conclusions.
13:20 - 13:55 Birte Forstmann and Brandon Turner “Model-based cognitive neuroscience”
Abstract
Cognitive neuroscientists study how the brain implements particular cognitive processes such as perception, learning, and decision- making. Traditional approaches in which experiments are designed to target a specific cognitive process have been supplemented by two recent innovations. First, formal cognitive models can decompose observed behavioral data into multiple latent cognitive processes, allowing brain measurements to be associated with a particular cognitive process more precisely and more confidently. Second, cognitive neuroscience can provide additional data to inform the development of formal cognitive models, providing greater constraint than behavioral data alone. We argue that these fields are mutually dependent; not only can models guide neuroscientific endeavors, but understanding neural mechanisms can provide key insights into formal models of cognition.
13:55 - 14:30 Jian Li “Reinforcement learning models and integration with fMRI”
Abstract
Reinforcement learning (RL) has witnessed its wide application in cognitive sciences in the past decades. In this session I will briefly introduce different RL models used in cognitive neuroscience/psychology research and how they can be integrated with fMRI techniques to better understand the computations that brain carries out during learning and inference.
14:30 - 14:45 Break
14:45 - 15:20 Jean Daunizeau “Model-based assessment of brain-behavior relationships”
Abstract
Functional outcomes (e.g., subjective percepts, emotions, memory retrievals, decisions, etc...) are partly determined by external stimuli and/or cues. But they may also be strongly influenced by (trial-by-trial) uncontrolled variations in brain responses to incoming information. In turn, this variability may provide critical information regarding how behaviourally relevant inputs are eventually transformed into functional outcomes. Assessing brain-behavior relationships thus requires considering the (possibly nonlinear and stochastic) impact of biological constraints of input-output transformations in the brain. In this talk, I will review the portfolio of existing approaches to decomposing the brain's transformation of stimuli into behavioural outcomes, in terms of the relative contribution of brain regions and their connections. In particular, I will highlight three novel techniques, namely: mass mediation analysis, artificial neural network modelling, and behavioral DCM (dynamic causal modelling). The aim here is for attendees to understand the strengths and weaknesses of each approach in turn, as well as gain practical know-how regarding how to perform such analyses.
15:20 - 16:10 Brandon Turner Hands on session: “A tutorial on joint models of neural and behavioral data”
Abstract
A growing synergy between the fields of cognitive neuroscience and mathematical psychology has sparked the development of several unique statistical approaches exploiting the benefits of both disciplines (Turner, Forstmann et al., 2017). One approach in particular, called joint modeling, attempts to model the covariation between the parameters of "submodels" intended to capture important patterns in each stream of data. Joint models present an interesting opportunity to transcend conventional levels of analyses (e.g., Marr’s hierarchy; Marr, 1982) by providing fully integrative models (Love, 2015). In this talk, we provide a tutorial of two flavors of joint models — the Directed and Covariance approaches. Computational procedures have been developed to apply these approaches to a number of cognitive tasks, yet neither have been made accessible to a wider audience. Here, we provide a step-by-step walkthrough on how to develop submodels of each stream of data, as well as how to link the important model parameters to form one cohesive model. For convenience, we provide code that uses the Just Another Gibbs Sampler (Plummer, 2003) software to perform estimation of the model parameters.
This session will be based on Palestro et al 2018 where the codes are at Github .
Please install R, JAGS, and the Wiener module for JAGS to follow the practical session on your computer.
After downloading and installing the most recent version of JAGS at http://mcmc-jags.sourceforge.net/ that corresponds to the operating system installed on your computer, install “rjags” package in R console by typing install.packages("rjags") and
require("rjags").
16:10 - 16:20 Panel discussion: Join the event on https://gmp3.cnf.io/ and ask your questions!
Organizers

