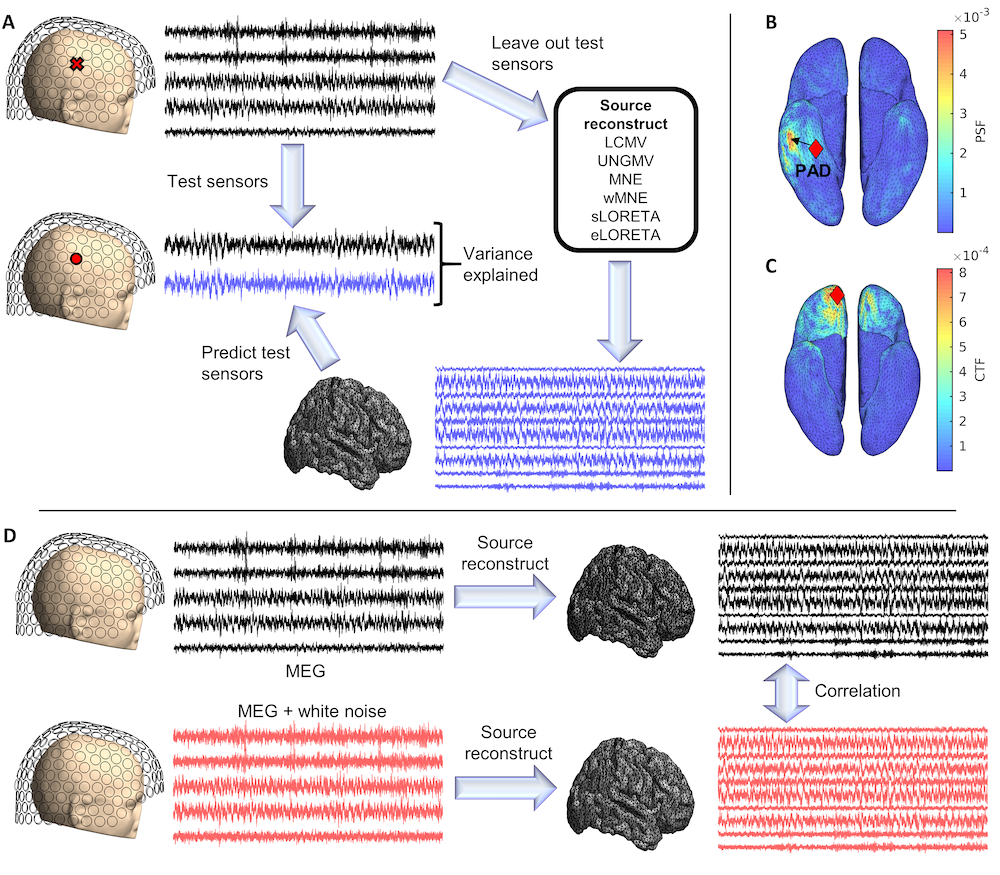
A new paper led by Luke Tait is published on Human Brain mapping. This study systematically evaluated the performance of six commonly-used source reconstruction algorithms. Using human resting-state MEG, we compared the algorithms using quantitative metrics, including resolution properties of inverse solutions and explained variance in sensor-level data. Next, we proposed a data-driven approach to reduce the atlas from the Human Connectome Project’s multi-modal parcellation of the human cortex based on metrics such as MEG signal-to-noise-ratio and resting-state functional connectivity gradients. This procedure produced a reduced cortical atlas with 230 regions, optimized to match the spatial resolution and the rank of MEG data from the current generation of MEG scanners.
Our results show that there is no “one size fits all” algorithm, and make recommendations on the appropriate algorithms depending on the data and aimed analyses. Our comprehensive comparisons and recommendations can serve as a guide for choosing appropriate methodologies in future studies of resting-state MEG.
The paper is now available online.